In a previous post entitled Genetics/DNA – part one, I stated that there are a few things we must address in regard to DNA before conclusions are made. The first has to do with the most noticeable structures called chromosomes. The second notable aspect of an organism’s genome has to do with coding and noncoding sequences. In this post, a third aspect of DNA will be covered and it has to do with the computer-like information stored in the rungs of the double helix called base pairs.
In a previous post entitled Genetics/DNA – part one, I stated that there are a few things we must address in regard to DNA before conclusions are made. The first has to do with the most noticeable structures called chromosomes. The second notable aspect of an organism’s genome has to do with coding and noncoding sequences. In this post, a third aspect of DNA will be covered and it has to do with the computer-like information stored in the rungs of the double helix called base pairs.
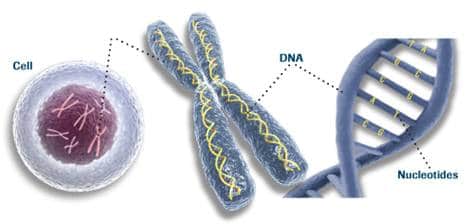
Each parent, in sexually reproducing organisms, passes along strands of information stored within DNA’s ladder-like rungs called polymers. New polymers are constructed by using half of each parent’s polymer, called a monomer. When two parental monomers are joined together, a new polymer or rung on the double helix ladder is created. This pairing is the basis of genetic code and these polymers are referred to as base pairs. Much like the binary language of computers that use the two variants of ones and zeros, genomes use five variants. DNA strands are sequenced using the letter representations GACT. The studying of these sequences is where evolutionary biologists have pieced similarities together.

In the binary language of computers, green is represented by 1-1-0. If one were to convert a photograph of a human and a chimpanzee side by side in a jungle into the binary language of computers, one would instantly see similarities. The binary code 1-1-0 for the color green would be quite prevalent, yet so would the code for black, which is 0-0-0. The code for green could accurately be describing organisms that produce chlorophyll, but no species of plant could be determined. Unfortunately, all non-plant, green organisms like leaf bugs and iguanas would also fall into the same descriptor of binary code 1-1-0. Continuing to view our jungle photo in binary code, black would also be dominantly repetitive, but what would it’s code represent? Perhaps both chimpanzee and human pupils would be linked to having a strong correlation, but binary code 0-0-0 would also be representational of lifeless shadows. As in the analogy of a photo converted to binary language, finding similar genetic sequences in various genomes of varying organisms also demonstrates to be inconsequential.

Sir Gavin de Beer was an evolutionary embryologist, Fellow of the British Royal Society, and served as Director of the British Museum of Natural History. He has done extensive research on the embryonic development between organisms that are visually and structurally homologous. His experiments were designed to trace the development of certain attributes from the moment of fertilization until full development. The results of his work revealed that fully formed corresponding homologies do not develop from the same embryonic location. Sir Gavin de Beer realized that similar attributes develop from a fertilized egg in different locations, depending on the species. This means that if homologous structures do not develop from the same embryonic location, then they are not genetically related because their development is not controlled by their homologous (similar) DNA. In short, embryonic homology promptly discounts any possible attempt of supporting evolution via genetic homology. Sir Gavin de Beer said it best when saying, “It is now clear that the pride with which it was assumed that the inheritance of homologous structures from a common ancestor explained homology was misplaced; for such inheritance cannot be ascribed to identity of genes.” If the work of De Beer revealed that homologous structures among differing species do not develop from the same embryonic location, then no genetic relationships can be drawn between two species with homologous sequences because their development is not controlled by their homologous DNA. Due to this fact, what significance is there in finding similar genetic sequences in the genome of varying species?
Another interesting aspect of base pairs represented in human and chimpanzee genomes has to do with numbers. The claim is that a 95% relational tie can be made due to the study of the genetic material in our DNA, yet what percentage does just the base pair information give us? Chimpanzees have 2.9 billion base pairs in their genome and we humans have 3.2 billion. For the sake of this argument, let’s say that the first 2.9 billion base pairs in our genome are a perfect match to that of chimpanzees. Without a single variant on any given polymer, there would still be 300 million differences between the two genomes. Humans have an extra 300 million base pairs. So, what would be our percentage of being genetically related to chimpanzees based solely on the first 2.9 billion polymers being a perfect match? 2.9 billion divided by 3.2 billion is equal to 0.90625, meaning that in this fictitiously error free scenario of comparing base pairs, we are approximately 90.6% related to chimpanzees. Adding in the potential mismatches, the thousands of differences in coding and noncoding sequences, and the fact that humans and chimpanzees have a different number of chromosomes, how scientific is it to claim a relational tie between these two organisms at all?
While we are on the subject of base pairs and the number we have in our genome compared to chimpanzees. Evolutionary speaking, it makes a lot of sense that we have a larger genome because we are “higher” lifeforms. Yet, is that really the case across all life?
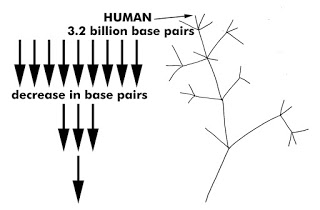
Before life began, 3.5-ish billion years ago, DNA was nonexistent or extremely rudimentary. This is why rudimentary organisms are placed toward the trunk of Darwin’s tree of life and highly evolved and complex organisms like humans are placed on the branches. There is an idea that complexity of life increases as one travels from the trunk to the branches. So, do we humans have the largest and most complex genome? It is larger than a chimpanzee’s, but is there a consistent increase in genetic complexity from trunk to branches on the Tree of Life? Do unicellular organisms have the least amount of base pairs in their genomes? According to Darwin’s Tree of Life, there should be a consistent increase in genetic complexity as organisms move from
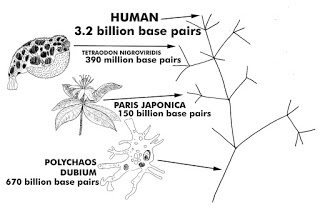
rudimentary or “lower” to “higher” lifeforms, yet what does the data show? The sequencing of various species has revealed their genomes to be all over the board. There is no consistent genetic data that verifies the idea of “lower” to “higher” or rudimentary to complex. Some unicellular organisms like the Polychaos Dubium have 670 billion base pairs which is 200 times larger than ours. Yet, a puffer-fish which is drastically more complex than a unicellular organism or a flower, has only a mere 390 million base pairs. How accurate does the Tree of Life seem in light of empirical data found in various lifeforms’ genomes?
What genetic data supports relational ties between species? What genetic data supports the idea of descent with modification from a common ancestor? What data supports the idea that complex DNA evolved from less complex DNA? What aspect of the field of genetics has generated empirical data that verifies evolution?In my book What Is Evolution?, I dive deep into these questions and many others that need to be asked. I hope you will join me.
 In a previous post entitled Genetics/DNA – part one, I stated that there are a few things we must address in regard to DNA before conclusions are made. The first has to do with the most noticeable structures called chromosomes. The second notable aspect of an organism’s genome has to do with coding and noncoding sequences. In this post, a third aspect of DNA will be covered and it has to do with the computer-like information stored in the rungs of the double helix called base pairs.
In a previous post entitled Genetics/DNA – part one, I stated that there are a few things we must address in regard to DNA before conclusions are made. The first has to do with the most noticeable structures called chromosomes. The second notable aspect of an organism’s genome has to do with coding and noncoding sequences. In this post, a third aspect of DNA will be covered and it has to do with the computer-like information stored in the rungs of the double helix called base pairs. Each parent, in sexually reproducing organisms, passes along strands of information stored within DNA’s ladder-like rungs called polymers. New polymers are constructed by using half of each parent’s polymer, called a monomer. When two parental monomers are joined together, a new polymer or rung on the double helix ladder is created. This pairing is the basis of genetic code and these polymers are referred to as base pairs. Much like the binary language of computers that use the two variants of ones and zeros, genomes use five variants. DNA strands are sequenced using the letter representations GACT. The studying of these sequences is where evolutionary biologists have pieced similarities together.
Each parent, in sexually reproducing organisms, passes along strands of information stored within DNA’s ladder-like rungs called polymers. New polymers are constructed by using half of each parent’s polymer, called a monomer. When two parental monomers are joined together, a new polymer or rung on the double helix ladder is created. This pairing is the basis of genetic code and these polymers are referred to as base pairs. Much like the binary language of computers that use the two variants of ones and zeros, genomes use five variants. DNA strands are sequenced using the letter representations GACT. The studying of these sequences is where evolutionary biologists have pieced similarities together. In the binary language of computers, green is represented by 1-1-0. If one were to convert a photograph of a human and a chimpanzee side by side in a jungle into the binary language of computers, one would instantly see similarities. The binary code 1-1-0 for the color green would be quite prevalent, yet so would the code for black, which is 0-0-0. The code for green could accurately be describing organisms that produce chlorophyll, but no species of plant could be determined. Unfortunately, all non-plant, green organisms like leaf bugs and iguanas would also fall into the same descriptor of binary code 1-1-0. Continuing to view our jungle photo in binary code, black would also be dominantly repetitive, but what would it’s code represent? Perhaps both chimpanzee and human pupils would be linked to having a strong correlation, but binary code 0-0-0 would also be representational of lifeless shadows. As in the analogy of a photo converted to binary language, finding similar genetic sequences in various genomes of varying organisms also demonstrates to be inconsequential.
In the binary language of computers, green is represented by 1-1-0. If one were to convert a photograph of a human and a chimpanzee side by side in a jungle into the binary language of computers, one would instantly see similarities. The binary code 1-1-0 for the color green would be quite prevalent, yet so would the code for black, which is 0-0-0. The code for green could accurately be describing organisms that produce chlorophyll, but no species of plant could be determined. Unfortunately, all non-plant, green organisms like leaf bugs and iguanas would also fall into the same descriptor of binary code 1-1-0. Continuing to view our jungle photo in binary code, black would also be dominantly repetitive, but what would it’s code represent? Perhaps both chimpanzee and human pupils would be linked to having a strong correlation, but binary code 0-0-0 would also be representational of lifeless shadows. As in the analogy of a photo converted to binary language, finding similar genetic sequences in various genomes of varying organisms also demonstrates to be inconsequential.